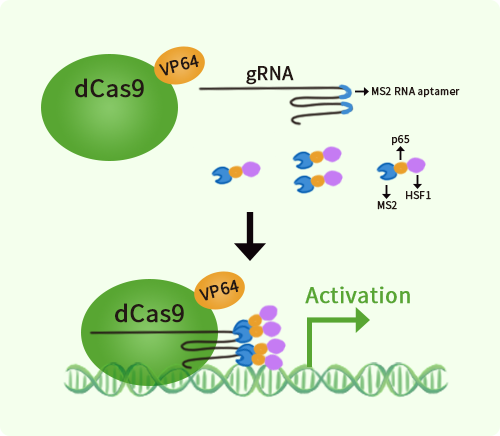
Mouse Ddx11 activation kit by CRISPRa
CAT#: GA216449
Ddx11 CRISPRa kit - CRISPR gene activation of mouse DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11
CNY 12,255.00
Specifications
| Product Data | |
| Format | 3 gRNAs (5ug each), 1 scramble ctrl (10ug) and 1 enhancer vector (10ug) |
| Symbol | Ddx11 |
| Locus ID | 320209 |
| Kit Components | GA216449G1, Ddx11 gRNA vector 1 in pCas-Guide-GFP-CRISPRa GA216449G2, Ddx11 gRNA vector 2 in pCas-Guide-GFP-CRISPRa GA216449G3, Ddx11 gRNA vector 3 in pCas-Guide-GFP-CRISPRa 1 CRISPRa-Enhancer vector, SKU GE100056 1 CRISPRa scramble vector, SKU GE100077 |
| Disclaimer | These products are manufactured and supplied by OriGene under license from ERS. The kit is designed based on the best knowledge of CRISPRa SAM technology. The efficiency of the activation can be affected by many factors, including nucleosome occupancy status, chromatin structure and the gene expression level of the target, etc. |
| Reference Data | |
| RefSeq | NM_001003919, NM_001348292 |
| Synonyms | 4732462I11Rik; CHL1; CHLR1; essa15a; KRG2 |
| Summary | DNA-dependent ATPase and ATP-dependent DNA helicase that participates in various functions in genomic stability, including DNA replication, DNA repair and heterochromatin organization as well as in ribosomal RNA synthesis. Its double-stranded DNA helicase activity requires either a minimal 5'-single-stranded tail length of approximately 15 nt (flap substrates) or 10 nt length single-stranded gapped DNA substrates of a partial duplex DNA structure for helicase loading and translocation along DNA in a 5' to 3' direction. The helicase activity is capable of displacing duplex regions up to 100 bp, which can be extended up to 500 bp by the replication protein A (RPA) or the cohesion CTF18-replication factor C (Ctf18-RFC) complex activities. Shows also ATPase- and helicase activities on substrates that mimic key DNA intermediates of replication, repair and homologous recombination reactions, including forked duplex, anti-parallel G-quadruplex and three-stranded D-loop DNA molecules. Plays a role in DNA double-strand break (DSB) repair at the DNA replication fork during DNA replication recovery from DNA damage. Recruited with TIMELESS factor upon DNA-replication stress response at DNA replication fork to preserve replication fork progression, and hence ensure DNA replication fidelity (By similarity). Cooperates also with TIMELESS factor during DNA replication to regulate proper sister chromatid cohesion and mitotic chromosome segregation (PubMed:17611414). Stimulates 5'-single-stranded DNA flap endonuclease activity of FEN1 in an ATP- and helicase-independent manner; and hence it may contribute in Okazaki fragment processing at DNA replication fork during lagging strand DNA synthesis. Its ability to function at DNA replication fork is modulated by its binding to long non-coding RNA (lncRNA) cohesion regulator non-coding RNA DDX11-AS1/CONCR, which is able to increase both DDX11 ATPase activity and binding to DNA replicating regions (By similarity). Plays also a role in heterochromatin organization (PubMed:21854770). Involved in rRNA transcription activation through binding to active hypomethylated rDNA gene loci by recruiting UBTF and the RNA polymerase Pol I transcriptional machinery (By similarity). Plays a role in embryonic development and prevention of aneuploidy (PubMed:17611414). Involved in melanoma cell proliferation and survival. Associates with chromatin at DNA replication fork regions. Binds to single- and double-stranded DNAs (By similarity).[UniProtKB/Swiss-Prot Function] |
Documents
Resources
| 基因表达相关资源 |
Other Versions
| SKU | Description | Size | Price |
|---|---|---|---|
| KN504377 | Ddx11 - KN2.0, Mouse gene knockout kit via CRISPR, non-homology mediated. |
CNY 8,680.00 |


 United States
United States
 Germany
Germany
 Japan
Japan
 United Kingdom
United Kingdom
 China
China